Chapter 1 Familiarization
1.1 R as a calculator
Assuming that you have installed and started R as in the last section, let’s start to use R as a simple calculator. In the console (bottom left of the screen), run the following by typing each line and hitting enter.
# Some simple addition
2+2## [1] 4R can, of course, do everything that a calculator can do.
6*8## [1] 48100/10## [1] 104^3 # This is 4 to the power of 3. ## [1] 64R has all the usual mathematical functions. For example, trigonometric functions such as sin(), cos(), are available, as are the
exponential and log functions exp(), log(). The absolute value is
given by abs(), and round() will round a value to the nearest
integer.
pi # the constant 3.14159265...## [1] 3.141593floor(pi) # round down to the nearest integer## [1] 3ceiling(pi) # round up to the nearest integer## [1] 4sin(0)## [1] 0exp(1) # exp() is the exponential function## [1] 2.718282log(5) # unless you specify the base, R will assume base e## [1] 1.609438log(5, base=10) # base 10## [1] 0.69897Some parameters we pass to a function (these are called arguments) are
mandatory, and some are optional. Arguments are separated by
a comma. As you see above, log() requires at least
one argument, which is the number(s) to take the log of. However, the
base argument is optional. If we don’t specify a base, R
uses a default value. We can see that R will default to using natural logarithms (base
\(e\)) by looking at the help page (by typing help(log) or ?log at the
command prompt).
The order of the arguments can be important in function, but to avoide confusion arguments can be named, such as for the log() function which has arguments
log(x, base=exp(1)). If I specify which arguments are which using the
named values, then order doesn’t matter.
# Demonstrating order does not matter if you specify
# which argument is which
log(x=5, base=10) ## [1] 0.69897log(base=10, x=5)## [1] 0.69897If we don’t specify which argument is which, R will decide that x
is the first argument, and base is the second.
# If not specified, R will assume the second value is the base...
log(5, 10)## [1] 0.69897log(10, 5)## [1] 1.430677Of course, functions can be combined- R Studio will help you, but try to make sure that the brackets are in the right place and match.
sin(log(42))## [1] -0.56140031.2 Reproducability and saving your work
R is a script based language, and there isn’t (for most things) a point-and-click interface like in excel, for example. One great benefit of R is that writing scripts leaves a clear description of exactly what steps were performed, and helps to make sure that that someone else can see how you got your results. This reproducibility is a critical aspect of sharing your methods and results with other students, colleagues, and the world at-large.
1.2.1 Working within an R Script File
The first step in any new analysis or project is to create a new R
Script file. This can be done by selecting the
File -> New File -> R Script
Once you’ve created a new R Script file, you’ll be presented with four different panes that you can interact with.
| Pane | Location | Description |
|---|---|---|
| Editor | Top Left | Where you edit the script. This is where you should write almost all of your R code. You should also execute your code from this pane. Because nobody writes code correctly the first time, you’ll inevitably make some change, and then execute the code again. This will be repeated until the code finally does what you want. |
| Console | Bottom Left | You can execute code directly in this pane, but the code you write won’t be saved. I recommend only writing stuff here if you don’t want to keep it. I only type commands in the console when using R as a calculator and I don’t want to refer to the result ever again. |
| Environment | Top Right | This displays the current objects that are available to you. |
| Miscellaneous | Bottom Right | This pane gives access to the help files, the files in your current working directory, and your plots (if you have it set up to show here.) |
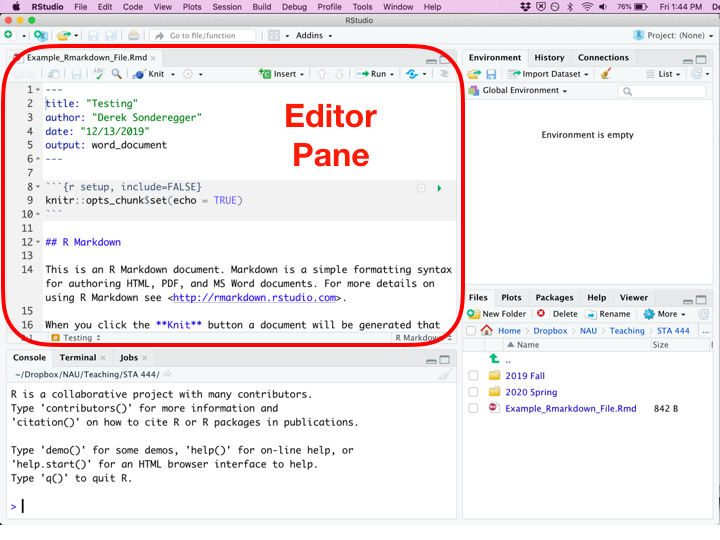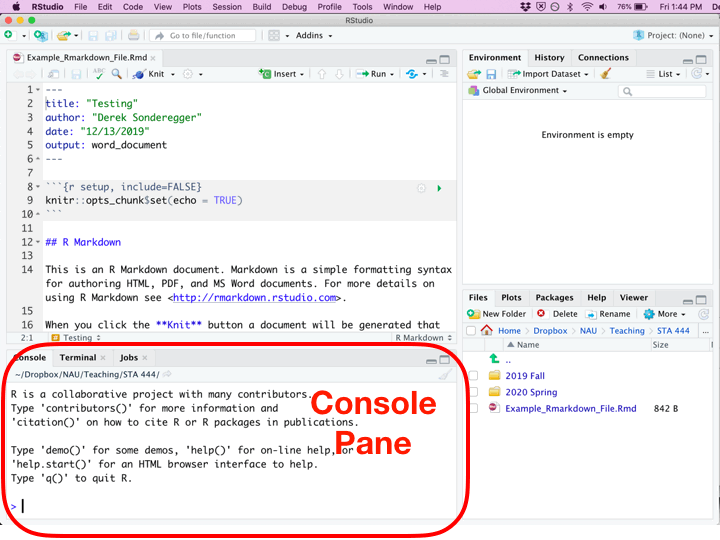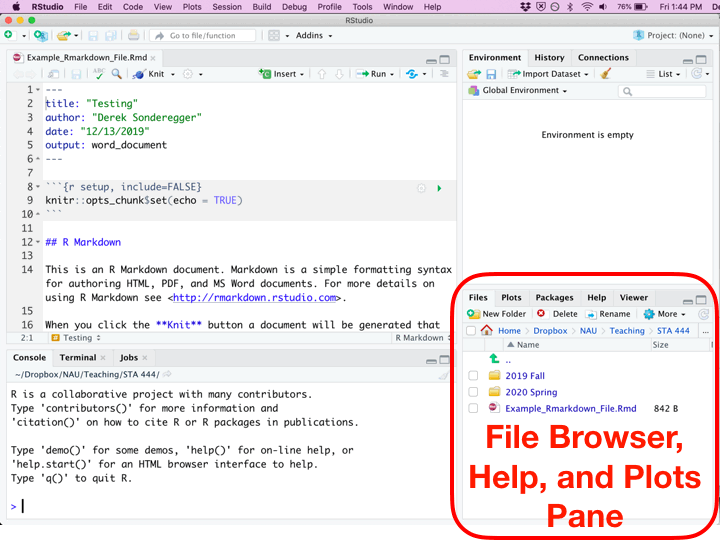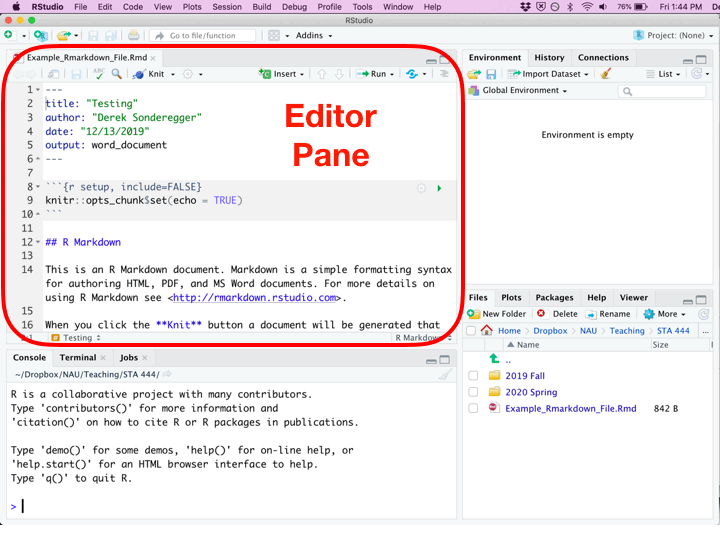
While writing an R Script file, each of the code chunks can be executed in a couple of different ways.
- Press the run key at the top of the pane to run the last line.
- Highlight the code that is wanted and run more than one line.
- Hit the “re-run” button next to the run button, which will re-run the last code executed. This is useful if you make a small change and want to re-run it.
1.2.3 Exercise: Setting up an R Script
Throughout this book, there will be various exercises: exercises are the best way to learn to do something, so I encourage you to have a go. Here is the first!
Let’s suppose we want to simulate rolling a six-sided dice one hundred
times. We can do this using the sample command.
sample(6,size=100,replace=TRUE)- Open a new R script.
- Copy this command into an R script and run it
- Now suppose we want to take the mean of this data. We can do this by putting the function mean() around the current command. Make the change and run the command.
- What happens if we now have 1000 dice? What if we have 10000? Do we get the mean you expect? (Hint: What is the mean of a discrete uniform[1,6] distribution)
1.3 Assigning variables
We will want to assign a variable to take some value. We do this using an arrow <-. (This is two keystrokes on the keyboard! )
tau <- 2*pi # create two variables
my.test.var <- 42 # notice they show up in 'Environment' tab in RStudio!
tau## [1] 6.283185my.test.var## [1] 42tau * my.test.var## [1] 263.8938R actually allows assignment using either an arrow <- or an equals sign
=. While R supports both, I suggest using the arrow <- .
tau <-2*pi
log(5, base=10) # base 10## [1] 0.69897tau = 2* pi # will work
log(5,base<-10) # May work but not reccommended## [1] 0.69897When specifying arguments for functions, we should use the name=value
notation. You might be tempted to use the <- notation with functions; don’t do that as the name=value notation is making an association
mapping and not a permanent assignment. In summary, I recommend using
<- for permanent assignment and = for temporarily assigning values in functions.
Variable names cannot start with a number, may not include spaces, and are case sensitive: Foo and foo are two different variable names, so being
consistent in your capitalization scheme is quite helpful. I try to use the “camelcase” convention where I write my variables as myVariableName.
You can see the name of any variables you have assigned in the environment tab in the environment pane (top-right). We can remind ourselves how we spelled a variable name and capitalization, and see a preview of what is stored in the variable.
1.3.1 Vectors
We often want to deal with a collection of data, and the most fundamental collection of values is called a vector in R. Vectors must always be of the
same type (e.g. all integers or all character strings). To create a
vector, we use the collection function c().
x <- c('Bingo Little','Roderick Spode','Aunt Agatha','Madeline Bassett')
x## [1] "Bingo Little" "Roderick Spode" "Aunt Agatha" "Madeline Bassett"y <- c( 3, 1, 4, 5 )
y## [1] 3 1 4 5It is common to have to make sequences of integers, and R has a
shortcut to do this. The notation A:B will produce a vector starting
with A and incrementing by one until we get to B.
2:6## [1] 2 3 4 5 6Nearly every function in R behaves correctly when being given a vector of values.
x <- c(4,7,5,2) # Make a vector with four values
log(x) # calculate the log of each value.## [1] 1.3862944 1.9459101 1.6094379 0.6931472x+x## [1] 8 14 10 410*x## [1] 40 70 50 201.4 Packages
One of the greatest strengths about R is that so many people have
developed free add-on packages to do some useful additional task. For
example, readxl is a package that allows R to read and write excel files. It’s not loaded when R is first started, but if you want to work with excel files, you can add it on simply.
We need to do a two-step procedure to use a package: 1. Install the package 2. Load the package
Step 1 is sometimes slow, but we only need to do this once, unless we change computers. Step 2 is normally fairly rapid.
We’ll demonstrate using the cowsay package, which is a stupid but fun package.
To download and install the package from the default site, the Comprehensive R Archive Network (CRAN), you just need
to ask RStudio it to install it via the menu Tools ->
Install Packages.... Once there, you just need to give the name of the
package (cowsay) and RStudio will download and install the package on
your computer.
Once a package is downloaded and installed on your computer, it is available, but is not loaded into your current R session by default. The reason it isn’t loaded is that there are thousands of packages, some of which are quite large and only used occasionally. So to improve overall performance only a few packages are loaded by default and the you must explicitly load packages whenever you want to use them. You only need to load them once per session/script.
Let’s try our cowsay package. Remember to install it first, then run the following
library(cowsay) # load the cowsay library
say("Hello World")##
## --------------
## Hello World
## --------------
## \
## \
## \
## |\___/|
## ==) ^Y^ (==
## \ ^ /
## )=*=(
## / \
## | |
## /| | | |\
## \| | |_|/\
## jgs //_// ___/
## \_)
## For a similar performance reason, many packages do not automatically load their datasets unless explicitly asked. Therefore when loading datasets from a package, you might need to do a two-step process of loading the package and then loading the dataset.
library(faraway) # load the package into memory
data("butterfat") # load the dataset into memory(This package faraway is a good example of a use of a package, as it contains a lot of data sets from books by Julian Faraway)
If you don’t need to load any functions from a package and you just want the data sets, you can do it in one step.
data('butterfat', package='faraway') # just load the dataset, not anything else
head(butterfat) # print out the first 6 rows of the data## Butterfat Breed Age
## 1 3.74 Ayrshire Mature
## 2 4.01 Ayrshire 2year
## 3 3.77 Ayrshire Mature
## 4 3.78 Ayrshire 2year
## 5 4.10 Ayrshire Mature
## 6 4.06 Ayrshire 2yearSimilarly, if I am not using many functions from a package, I might call
the functions using the notation package::function(). This is
particularly important when two packages both have functions with the
same name and it gets confusing which function you want to use. For
example the packages mosaic and dplyr both have a function tally.
So if I’ve already loaded the dplyr package but want to use the
mosaic::tally() function I would use the following:
mosaic::tally( c(0,0,0,1,1,1,1,2) )## Registered S3 method overwritten by 'mosaic':
## method from
## fortify.SpatialPolygonsDataFrame ggplot2## X
## 0 1 2
## 3 4 1Finally, many researchers and programmers host their packages on GitHub
(or equivalent site) and those packages can easily downloaded using
tools from the devtools package, which itself can be downloaded from CRAN.
devtools::install_github("tidyverse/readxl")1.5 Finding Help
There are thousands of packages in R, and it’s not expected that anyone can know every R command- even after lots of experience, you will need to remind yourself how to do something. The good news is that there are lots of ways to get help.
1.5.1 How does this function work?
Every function comes with a help file, and generally this documentation is well written. Let’s suppose I am interested in how the rep function works. We can access the rep help page by searching in the help window (bottom right pane) or we can get the same page from the console by typing help(rep). This document shows what arguments the function expects and what it will return. At the bottom of the help page is often a set of
examples demonstrating different ways to use the function. As you learn more R, these help files become quite handy, but initially they can be challenging to understand.
1.5.2 How does this package work?
Some package authors provide a “vignette”, which is generally a tutorial on how to get started with a package. Some experience or knowledge is required with R and the topic in question, but these are normally great places to start. Generally we find these by googling “R package XXXX” and that will lead the documentation on CRAN that gives a list of functions in the package, and sometimes a vignette. Some well-written packages have their own websites and are very well documented, and certainly those that we use in this course have a lot of examples.
1.5.3 How do I do XXX?
If you know what you want to do, but not how to do it, a great place for help is the question and answer site stackoverflow. As well as asking your instructor/lecturer/friends how to do something, kind-hearted experts on the internet will provide help, and more usually someone will have asked the same question as you. However, don’t just cut-and-paste code from the Internet- think about why a particular chunk of code works, and you’ll learn more quickly.
1.6 Exercises
Create an RMarkdown file that solves the following exercises.
Calculate \(\log\left(6.2\right)\) first using base \(e\) and second using base \(10\). To figure out how to do different bases, it might be helpful to look at the help page for the
logfunction.Calculate the square root of 2 and save the result as the variable named sqrt2. Have R display the decimal value of \(\sqrt{2}\). Hint: use Google to find the square root function. Perhaps search on the keywords “R square root function”.
The
repfunction allows you to repeat one or more digits. Read the help file on `rep``to see how it works, paying particular attention to the examples at the bottom.
- Use the rep function to print out the digit “7” 15 times as follows:
## [1] 7 7 7 7 7 7 7 7 7 7 7 7 7 7 7- Use the rep function to repeat the digits “1,2,3” ten times.
## [1] 1 2 3 1 2 3 1 2 3 1 2 3 1 2 3 1 2 3 1 2 3 1 2 3 1 2 3 1 2 3This exercise walks you through installing a package with all the datasets collected by Julian Faraway.
- Install the package
farawayon your computer using RStudio. - Load the package using the
library()command. - Print out the dataset
broccoli. - Use the help file to describe what the columns of the dataset mean
- Install the package
1.2.2 Comment your code!
One tip for any coding language, including R, is to comment your code. This is to allow anyone else reading it, most probably a future version of yourself, to understand what you did and why. In R, a # will mean that the rest of the line is ignored.
You can save your script file using the disk button in the toolbars, or via the “File->Save (As)” menu. I would recommend you do this for this course, and have a different script for each chapter. You can copy and paste code from these instructions into your file- hover over each segment and select the “copy to clipboard” icon that appears.