For more information about Grid storage and explanation of the acronyms, see sections 1 and 2 of MICE Note 247. The file-naming scheme is defined in MICE Note 264.
For this exercise, assume that you want to get hold of the online monitoring histograms for run 1161.
This option is no longer available.
As an interim step while people learn to use the Grid directly, some of the data will be made available via a web-browser interface,
though you will still need your certificate in the browser.
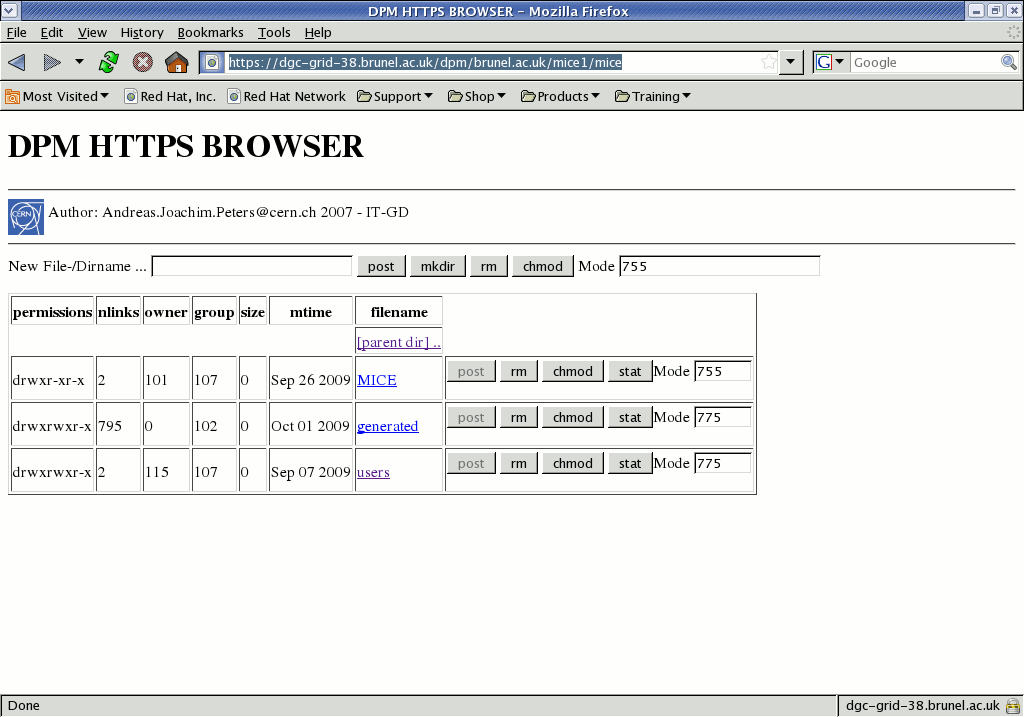
For online histograms, you can access data with a web browser via the https-enabled SE at Brunel:
https://dgc-grid-38.brunel.ac.uk/dpm/brunel.ac.uk/mice1/miceYou must have your Grid certificate in the browser (if not you will get an "Internal Server Error"); and you must be registered in the MICE VOMS server else you will get a cryptic message involving msg="No virtual ID mapping for <your DN>" If your browser pops up a message about not trusting the site, you have not yet imported the UK eScience CA root certificates and CRL.
(If you get a message about mixed secure and insecure content, it doesn't matter what you answer - it's that CERN logo in the corner. I'm working on it...)
The directory called MICE in the top row of the browser table is equivalent to that at the top of the LFC tree, so that we can easily see the path to follow: MICE -> Step1 -> 01100, to get to
https://dgc-grid-38.brunel.ac.uk/dpm/brunel.ac.uk/mice1/mice/MICE/Step1/01100/and scroll down to the file you want
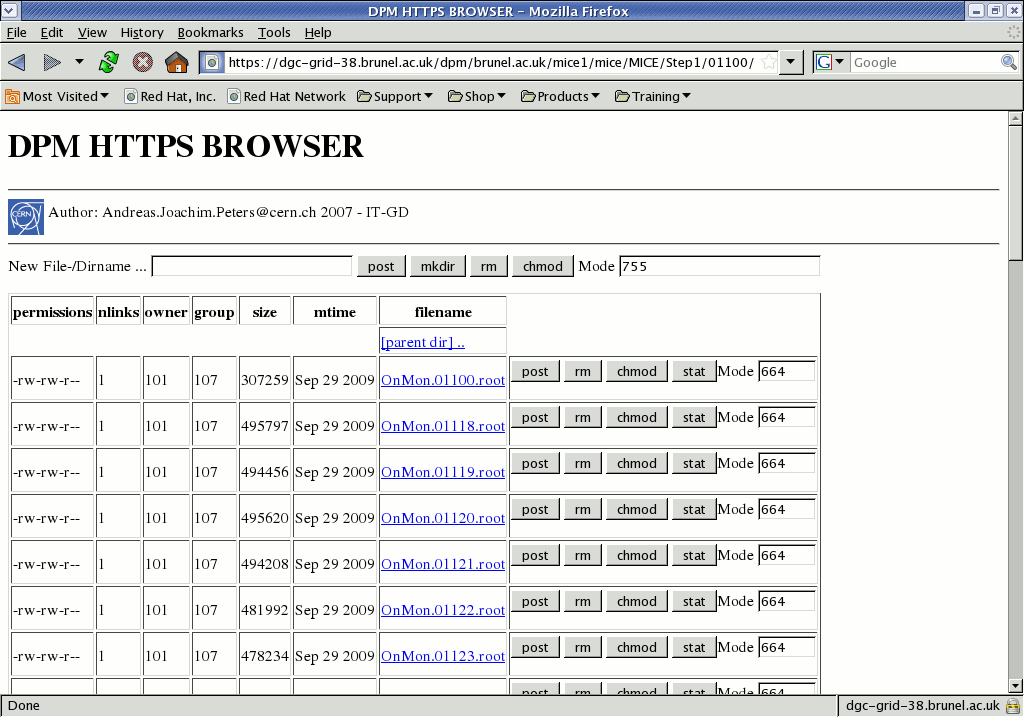
The browser shows the filesize and upload date, and the name itself provides a link to a SURL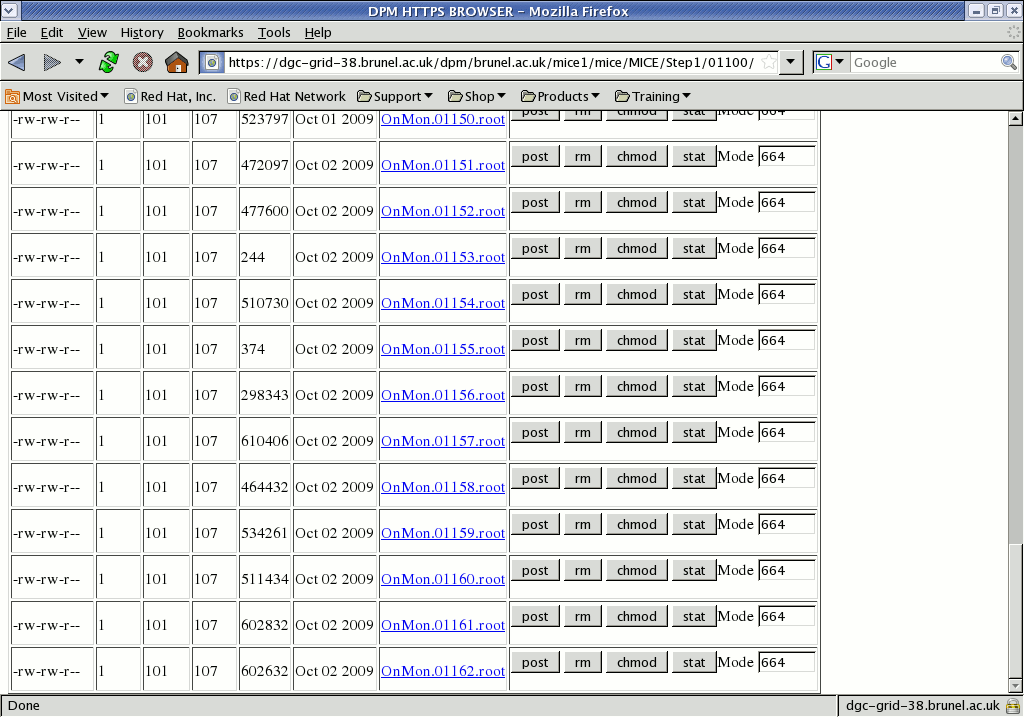
https://dgc-grid-38.brunel.ac.uk/dpm/brunel.ac.uk/mice1/mice/MICE/Step1/01100/OnMon.01161.rootthat can be used to access the file; this can be bookmarked or linked from a webpage (despite appearances, it is NOT actually a link to the file itself - see below).
To download the file click on the filename. Behind the scenes, this will redirect the browser to a different server (possibly on a different machine) that will provide the actual file itself. Note that the storage doesn't have any inherent understanding of what a "root file" is, so your browser is now pointing at something (a TURL) that is a lump of binary data with a weird filename.
Depending on your browser, you may be able to rename it or choose where to save it locally: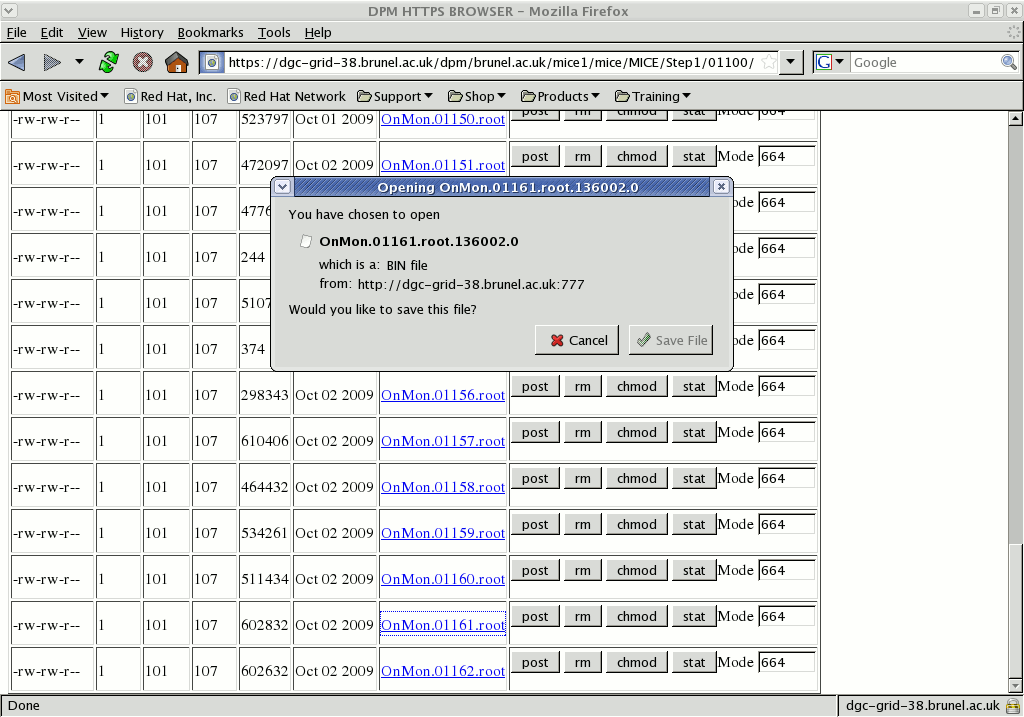
I now have the file on my desktop.
My laptop doesn't have ROOT on it, so...phsrjjn@turtle:~ > ls Desktop/ OnMon.01161.root.136002.0
at least I can check it arrived in one piece.phsrjjn@turtle:~ > md5sum Desktop/OnMon.01161.root.136002.0 0bc80c4fe7f5c6d6269c380d61914c4e Desktop/OnMon.01161.root.136002.0
The wget HTTP command-line client can authenticate using X.509 ("Globus-ified" or .pem format) certificates, and can thus be used to access data via this Web interface.
Note that the web server is a low-powered piece of hardware; please avoid encrypted transfers unless you have specific worries about data integrity. Also, if using CLI clients in scripts, please download files serially rather than setting up parallel transfers
If you get the following error message when downloading and the URL in the browser begins http://dgc-grid-38.brunel.ac.uk:777:
then your access is being filtered through a web cache or proxy; start from https://dgc-grid-38.brunel.ac.uk:883/dpm/brunel.ac.uk/mice1/mice instead.Authorization Required This server could not verify that you are authorized to access the document requested. Either you supplied the wrong credentials (e.g., bad password), or your browser doesn't understand how to supply the credentials required.
More information about the https front-end can be found at the DPM HTTPS wiki page.
Ultimately all the data will be available through the Grid, so start practising now! Grid clients imply a suitably configured gLite UI, and should be available on those EGEE resources that support the MICE VO for remote jobs.
(First, I don't yet have one, so I create today's proxy:
and some setupyoung: ~> voms-proxy-init -voms mice ... Creating proxy .................................. Done Your proxy is valid until Sat Oct 3 09:50:45 2009
)young: ~> setenv LFC_HOST lfc.gridpp.rl.ac.uk
The MICE data is arranged into a kind of virtualised filespace, inside the LFC. Lets look at the top of the MICE VO space:
That "MICE" is again the same MICE as above; it is the root of the "primary data" namespace. So a bit of common sense lets us jump straight to the LFN of the file we want:young: ~> lfc-ls /grid/mice Calibration Construction MICE TestBeam generated users
To download a file we simply use the lcg-cp command; this will track down the location of the real file we want and download it in a single swift move (but note the change in syntax):young: ~> lfc-ls /grid/mice/MICE/Step1/01100 OnMon.01100.root ... OnMon.01160.root OnMon.01161.root OnMon.01162.root
We're good to go...young: ~> lcg-cp --vo mice "lfn:/grid/mice/MICE/Step1/01100/OnMon.01161.root" "file:///tmp/OnMon.01161.root" young: ~> ls -l /tmp/OnMon.01161.root -rw-r--r-- 1 eesrjjn eesf 602832 Oct 2 22:09 /tmp/OnMon.01161.root
You may be wondering how to check that the file is uncorrupted. The Grid prefers a checksum algorithm called Adler32; this uses less CPU power than the common MD5 hash. The Grid clients include a crude Adler32 utility:
But how do you know the original value? The preferred route would be to have it in a database somewhere, but in the meantime it's embedded in the LFC comments field:young: ~> adler32 /tmp/OnMon.01161.root WARNING: SRM_PATH is defined, which might cause a wrong version of srm client to be executed WARNING: SRM_PATH=/opt/d-cache/srm ae82f954
You can also add a --checksum argument to lcg-cp, which will validate the transfer itself, but not any subsequent copies around your local systems.young: ~> lfc-ls --comment /grid/mice/MICE/Step1/01100/OnMon.01161.root /grid/mice/MICE/Step1/01100/OnMon.01161.root ae82f954
Note that online monitoring is currently stored on the Brunel server which is a low-powered piece of hardware, please avoid encrypted/checksummed transfers unless you have specific worries about data integrity. Also, if using CLI clients in scripts, please download files serially rather than setting up parallel transfers.