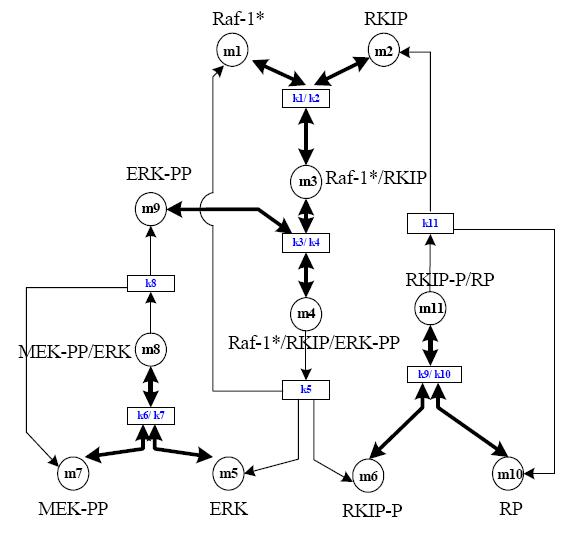
The RKIP pathway.
Raf-1, RKIP, ERK-PP, etc. are all proteins whose interactions are represented by this pathway. Raf-1/RKIP denotes a product of their interaction, where as the PP after MEK-PP or ERK-PP denotes the activated form of the protein. This means that ERK-PP is not in the strictest sense synonymous with ERK, however there is no standard way of writing this name so many researchers may write it as pERK or ERKPP.
Part 1
As discussed before, in this part we will use some of the on-line tools to familiarise ourselves with different aspects of this pathway and to gain full appreciation for the difficulties biologists may go through trying to find specific information using the standard information access tools.
Some of these resources may take a long time to load, in particular the Gene Ontology AmiGO tool, so please try to do some of these searches in parallel in different browser tabs/windows.
Spend roughly 10 minutes per question.
- Getting familiar with the pathway. In this exercise locate different components of the pathway and write down their synonyms. Try to understand the function of the pathway by looking at different species where it occurs and by following various links.
- Open the KEGG
site in one browser window. Looking at the whole MAPK pathway, can you
see where the RKIP pathway would fit on this network? Which of the
components from this network are also in the MAPK pathway? Click on the
components to find out more about them. Which pathways share these same
components?
- Open Entrez Gene in another
window search for RKIP. What did you notice about this particular
protein? Write down its
synonyms.
- For further information look on The Gene Ontology site, but beware that it takes a while to load.
- Reading about the pathway.
- Use PubMed and Web of Knowledge to do various searches about the pathway, using different kinds of keywords which you think would get you more information about the components or the pathway from literature.
- Using specialist websites.