MRes Bioinformatics: Systems Biology Lab 3: Biochemical Pathways
The aim of this lab is to give you practical experience in the construction and simulation of models of biochemical networks.
This lab refers to the slides for the lecture on
Modelling
dynamic behaviour.
BioNessie:
- BioNessie http://www.bionessie.com is a software tool which allows users to compose models of biochemical systems using biochemical equations, and to save these models as in SBML format (see http://www.sbml.org for model details on SBML---basically, it is a mark-up language for biochemical models).
- BioNessie enables the simulation of dynamic behaviour of biochemical models using a differential equation solver, and displays the results graphically. It also supports advanced analysis methods, including parameter scans and sensitivity analysis.
- BioNessie is available at
/Desktop/ZSBPS/bps/bin/bps.exe. For more information about using BioNessie, please see the BioNessie user manual.
Exercise 1
Use BioNessie to create a Mass Action based computational model for each of the following biochemical descriptions and produce simulations for each of the models.
Descriptions
Rate constants and initial concentrations required for the models are:
- Rate Constants: k1 = 1; k2 = 0.5; k3 = 2
- Initial Concentrations: [A] = 5; [B] = 1; [C] = 0; [D] = 0; [E] = 1; [AE]= 0
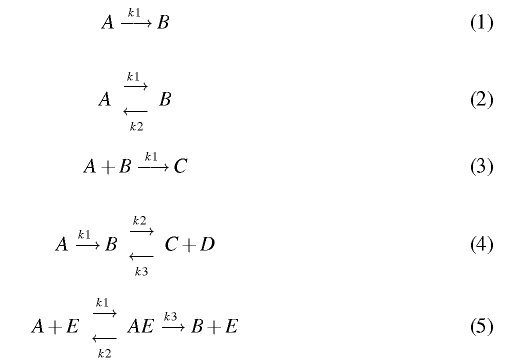
Exercise 2
(A) With the experience of modelling equation (5), can you model and
simulate (i) a Metabolic Pathway and (ii) Signalling Pathway? See slide
28 from the lecture notes for outlines of the pathways. (A more detailed
description of a 3-stage signalling pathway is on slide 50).
Refer to the following information to model and simulate the pathways.
- Rate constants are the same as that in equation (5).
- Initial concentration of all enzyme-substrate complexes is given to 0.
- Initial concentration of substrates, products and enzymes involved in these two pathways are:
- Metabolic Pathway: [E1] = [E2] = [E3] = 1; [S] = 100; [S'] = [S''] = [S'''] = 0
- Signalling Pathway: [Input Signal] = 1; [S1] = [S2] = [S3] = 100; [P1] = [P2] = [P3] = 0
(B) Create models for the following small pathways and investigate their
behaviours:
- phosphorylation-dephosphorylation stage, Mass-Action (slide 42, or 43 or
44).
- 2-stage phosphorylation cascade, Mass-Action (slide 48).
- 2-stage phosphorylation cascade, with negative feedback, Mass Action
(slide 52).
(C) Can you use BioNessie to create computational models for the pathways on slide 53 and slide 55? Michaelis-Menten kinetic laws will be applied for both pathways. Refer to Introduction - Enzyme Kinetics and the Michaelis-Menten equation for Michaelis-Menten kinetics. BioNessie provides users with the function of defining kinetic laws on their own. Refer to BioNessie user manual to find out how to create a user-defined kinetic law.
This page was last updated on 27 March 2007