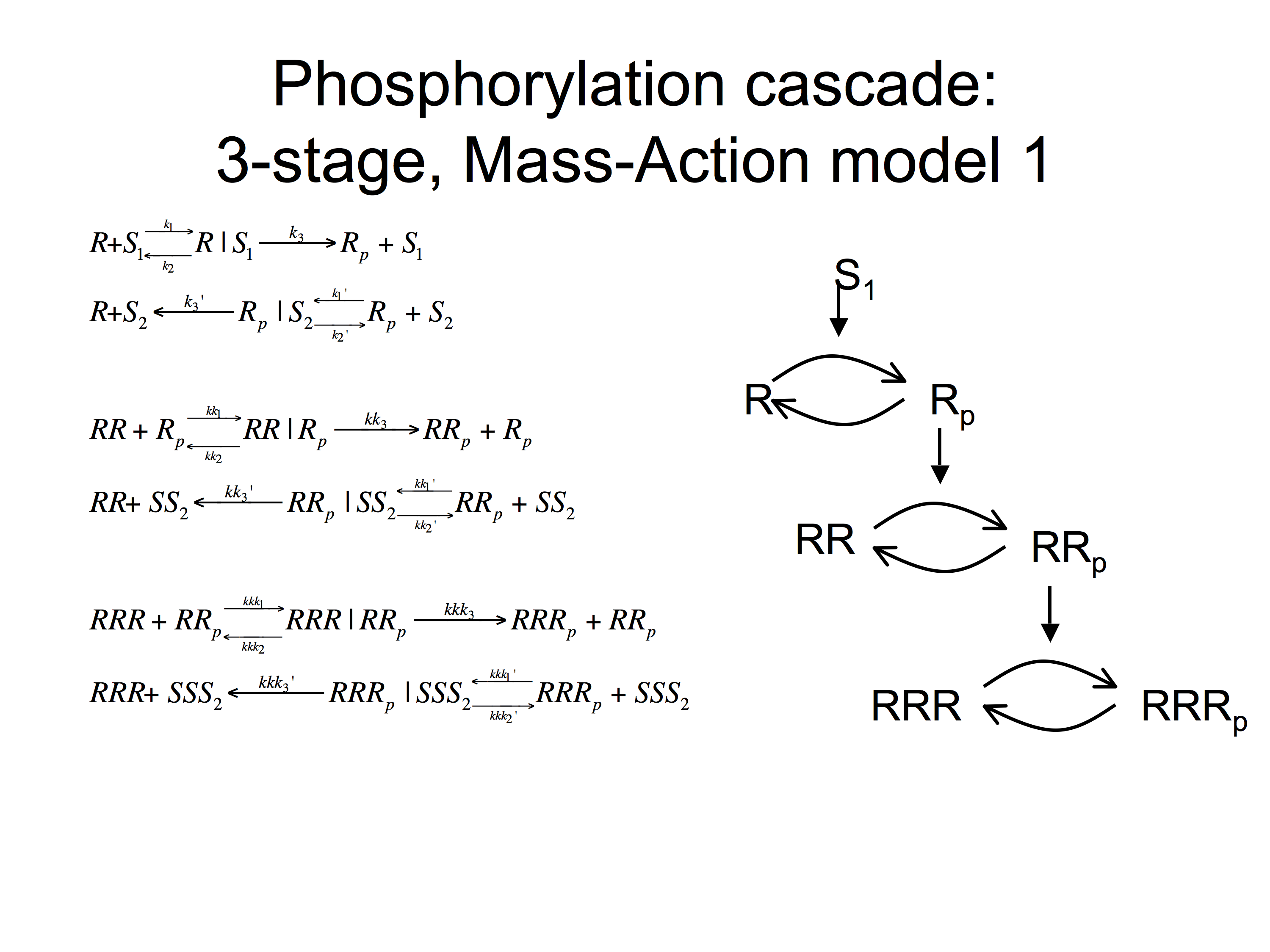
(1) Create an SBML model for the artificial signalling cascade (for outline form see below) [6 marks]
(2) Modify your model to include negative feedback [2 marks]
(2) Produce time-plot simulations for the behaviours of (1) and (2). Try to to produce oscillations for the negative feedback model. [4 marks]
(3) Give a representation of the pathway with negative feedback and explicitly including the phosphotases as a graph, using set notation. [4 marks]
(4) Give a representation of the pathway with negative feedback and explicitly including the phosphotases as an adjacency matrix [4 marks]
Notes:
Your submission should comprise
(i) a hard-copy submission including the
(ii) An email submission of
your SBML model and any accompanying simulation files.
PENALTY FOR NO ELECTRONIC SUBMISSION = -6 marks
to gux@dcs.gla.ac.uk
using as the subject line the following:
MRes systems biology coursework submission from YOURNAME

Rate constants and initial concentrations required for the modelling and simulation of the pathway are:
k1=100, k2=4, k3=1Take initial concentrations of the unphosphoyrlated proteins as 100 units, the kinases as 1 unit, the phosphotases as 1 unit, and everything else as zero.